One particular nice thing about SVR is that the weight of each feature reflects the feature’s true contribution. If some features are highly correlated (this is common in neuroimaging data – e.g. adjacent voxel activity are very similar), you expect that the corresponding weights to be similar. SVR exactly does that. Linear regression, on the other hand, will give big weight to the feature which is best correlated to the dependent variable (y). Here is a demonstration in MatLab.
In the following simulation, we have 10 features which are highly correlated (see figure below).
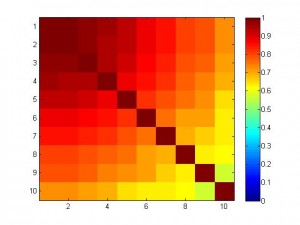
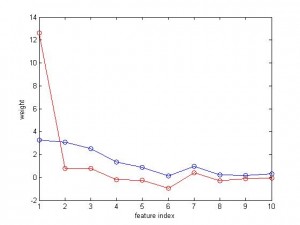
And the variable to be predicted is fairly nicely correlated with the features. Below I plotted the weight of 10 features. The blue curve is for SVR, and the red is for linear regression. As you can see, the weight for SVR is smooth and balanced, but the weight for regression is like ‘winner takes all’, the 1st feature has a big weight but others very small.
Here are the source code in MatLab. normalize.m can be found at https://www.alivelearn.net/?p=1083
N = 1000;
M = 1;
t = randn(N,1);
clear r
m = 10;%1:10:100;
for M = m
x = [t];
for ii=1:M-1
x = [x t+ii*randn(N,1)/10];
end
x = normalize(x);
% t1 = randn(N,1);
% t2 = randn(N,1);
% x = [t1 t2];
% y = 3*t1 - 5*t2;
y = 2*t + randn(N,1)/2 + 0;
% corrcoef([x y]);
%
% b= glmfit(x,y);
for ii = 1
for jj=1
tic;model = svmtrain(y(1:N/2),x(1:N/2,:),['-s 4 -t 0 -n ' num2str(ii/2) ' -c ' num2str(1)]);toc
tic;zz=svmpredict(y(N/2+1:end),x(N/2+1:end,:),model);toc
tmp = corrcoef(zz, y(N/2+1:end));
r(M) = tmp(2);
end
end
w = model.SVs' * model.sv_coef;
b = -model.rho;
b1 = [w]
% regression
b2 = glmfit(x(1:N/2,:), y(1:N/2,:));
yy = x(N/2+1:end,:)*b2(2:end) + b2(1);
b2 = b2(2:end)
sum((zz - y(N/2+1:end)).^2)
sum((yy - y(N/2+1:end)).^2)
end
figure('color','w');imagesc(corrcoef(x));colorbar
caxis([0 1])
figure('color','w');plot(b1,'o-');hold on;plot(b2,'o-r');xlabel('feature index');ylabel('weight')
return
hold on;plot(m, r, 'ro-');xlabel('# of dimension'); ylabel('r')
figure('color','w');plot(m, r, 'ro-');
figure('color','w');plot(x(1:N/2,:), y(1:N/2), 'b.');
hold on;plot(x(N/2+1:end,:), zz, 'r.');
xlabel('x')
ylabel('y')
legend({'training','test'})
%figure('color','w'); plot(zz, y(N/2+1:end), '.'); axis equal;axis square;
%figure('color','w'); plot(zz - y(N/2+1:end), '.')



Very interesting. I was running into a similar problem with regression and wonder what the underlying reason for the winner takes all phenomenon is. Do you know more about it?
Thanks,
Henning
Great article!
e-SVR seems also to work great on Binary Classification Problems like pedestrian detection (Better than SVC…). I am a bit confused since Regression is for continuous output whereas Classification is discrete-oriented. Could you give me some insights about that (SVR vs SVC) ?
Cheers,
Djébril