SVM is mostly commonly used for binary classifications. But one branch of SVM, SVM regression or SVR, is able to fit a continuous function to data. This is particularly useful when the predicted variable is continuous. Here I tried some very simple cases using libsvm matlab package:
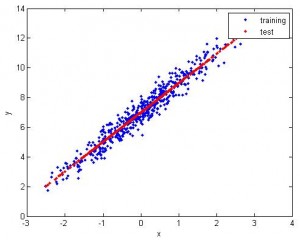
1. Feature 1D, use 1st half to train, 2nd half to test. The fitting is pretty good.
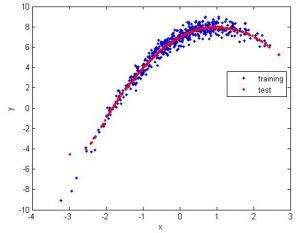
2. Still 1D, but apparently the data is nonlinear. So I use nonlinear SVR (radial basis). The fitting is good.
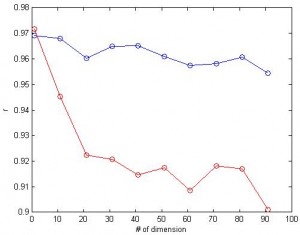
3. What if we have a lot of dimensions? Here I tried feature space with up to 100 dimensions and calculated the correlation between predicted values and the actual values. For linear SVR (blue), the number of dimension doesn’t affect the correlation much. (red: nonlinear, blue:linear, same data for both cases)
One property of SVR I like is that, when two features are similar (i.e. highly correlated), their weights are similar. This is in contrast with “winner take all” property of general linear model (GLM). This property is desired in brain imaging analysis: neighbor voxels have highly correlated signals and you want them to have similar weights.
About performance: If different features have different scales, then normalization of data will improve the speed of libsvm. Also, the cost parameter c also affects the speed. The larger c is, the slower libsvm is. For the simulated data I used, the parameters don’t affect the accuracy.
MatLab code: test_svr.m
The normalization function (copy and save it into normalize.m):
function data = normalize(d) % scale before svm % the data is normalized so that max is 1, and min is 0 data = (d -repmat(min(d,[],1),size(d,1),1))*spdiags(1./(max(d,[],1)-min(d,[],1))’,0,size(d,2),size(d,2));
libsvm: http://www.csie.ntu.edu.tw/~cjlin/libsvm/#matlab
options: -s svm_type : set type of SVM (default 0) 0 -- C-SVC 1 -- nu-SVC 2 -- one-class SVM 3 -- epsilon-SVR 4 -- nu-SVR -t kernel_type : set type of kernel function (default 2) 0 -- linear: u'*v 1 -- polynomial: (gamma*u'*v + coef0)^degree 2 -- radial basis function: exp(-gamma*|u-v|^2) 3 -- sigmoid: tanh(gamma*u'*v + coef0) -d degree : set degree in kernel function (default 3) -g gamma : set gamma in kernel function (default 1/num_features) -r coef0 : set coef0 in kernel function (default 0) -c cost : set the parameter C of C-SVC, epsilon-SVR, and nu-SVR (default 1) -n nu : set the parameter nu of nu-SVC, one-class SVM, and nu-SVR (default 0.5) -p epsilon : set the epsilon in loss function of epsilon-SVR (default 0.1) -m cachesize : set cache memory size in MB (default 100) -e epsilon : set tolerance of termination criterion (default 0.001) -h shrinking: whether to use the shrinking heuristics, 0 or 1 (default 1) -b probability_estimates: whether to train a SVC or SVR model for probability estimates, 0 or 1 (default 0) -wi weight: set the parameter C of class i to weight*C, for C-SVC (default 1) The k in the -g option means the number of attributes in the input data.
>



Your MatLab code goes wrong:
??? Error using ==> normalize at 43
A must be an ATOM.
Error in ==> test_svr at 13
x = normalize(x);
Looks like the normalize function is missing. Try:
function data = normalize(d)
% scale before svm
% the data is normalized so that max is 1, and min is 0
data = (d -repmat(min(d,[],1),size(d,1),1))*spdiags(1./(max(d,[],1)-min(d,[],1))’,0,size(d,2),size(d,2));
Would you please publish the data set, the liblinear SVR input parameters, and the output accuracy (MSE, R2) for the non-linear case. I would like to test this and am having some trouble getting the octave interface compiled and also understanding the SVR output
Thanks
Thank you very much for svr_test.m example with libsvm.
Excuse me,
a question about svr_test.m mfile.
Is it true that after the below commands zz and y(N/2+1:end)should be similar approximately? If it is true, why are zz and y(N/2+1:end)different?
tic;model = svmtrain(y(1:N/2),x(1:N/2,:),[‘-s 4 -t 2 -n ‘ num2str(ii/2) ‘ -c ‘ num2str(1)]);toc
tic;zz=svmpredict(y(N/2+1:end),x(N/2+1:end,:),model);toc
@Amin
No. zz is predicted value, and yy is original value. If the model is good, zz and yy is hopefully similar; otherwise they can be different.
Hi,
I have been working on epsilon SVR trying to figure out the best model, and I think I did based already. My question is, is it possible to obtain the regression coefficients/magnitudes for SV used in my model from the svmtrain code? If not, what is the best way to do it? I am mostly interested in finding something (SV magnitures/coef) like the regression coefficients from Multiple Linear regression … Is this possible?
Thanks in advance!
regards,
Are w and b in test_svr.m what you are looking for?
w = model.SVs’ * model.sv_coef
b = -model.rho
Thanks for the reply! Actually I am looking for the coef from training set I have. Can w and b be obtained from the svmtrain code on the original training set we have? My ultimate goal is to obtain the regression coef and use them to predict a different data set (not used in the original set). I hope it is more clear now.
w and b are from the training model and not from the test data.
hi
I want to know how we can apply error bound to our SVR to check the robustnees of my identified system?in epsilon regression
Hello
Could you tell me how can i use -p in SVR?does it have any difference with -e?
Hi Xu Cui..
I need your help about SVM Non Linear Reggression
could you please reply me in my email ?
Thanks
Hi Xu,
Great job about your tutorial regarding libsvm ! However, i don’t know how you get the plot above ? When i run your script I get only the red curve of the last plot. Could you help me to handle it ?
Keep up the hard work !
Thanks
Try change m=1.
It works thank you ! But what difference does that make between the different values of m ?
Increase dimension.
Thank you for your answer. By the way, do you think that we can extrapolate the function that interpolates the cloud to a wider range ?
I think extrapolation is dangerous – especially for nonlinear svr.
@Xu Cui
And do you have any idea how to do it ?
Thank you for your answers !
Hello
could you give some examples about precomputed kernel in SVR ?
how do we generate Xi and Xj ?
Hello
I’m working on svm regression for time series prediction. How can I find the opmtima C, e and gamma?
@mary
With the same technique used in regular SVM? Of course what to be maximized is the correlation of predicted vs actual, instead of accuracy.
Hey Xu
I am newbee to this field….just want to ask if we want to use 3 RBF functions how to do that in this case
Hi Xu,
as everyone else here, im working with SVR. Currently im working on the normalization data step and im using the normalization code you posted here. Yet i cant figure how to perform normalization for training and test data since training and testing data could have different min and max values. Should i use the min and max values of my training data to normalize the test data?
Thx in advance
@Guillermo
I think it is unfair to normalize training data and test data differently. So yes, I would use the min/max of the training data to scale the test data .
@Ahlad
Do you mean sigmoid? I never tried …
Dear Xu Cui,
I am very much interested in SVR. I am a beginner to SVM to guide me. I have a data set of mxn (100×40) with missing entries. I want to predict those missing entries through your code. But, right now the code uses 1D. can you please guide me on how should I alter ur code? Also, I want to know how to install the libsvm. I have learnt that SVM requires train set and test set. With my matrix how do i create a train and test set? Please guide me, i believe you can help me as soon as possible. Regards,manoraaju
@Mano
Mano,
After downloading libsvm (the matlab version) you simply add its path to MatLab’s path. Then you can use it.
Extending from 1D to 2D is very easy. My code above actually contains feature space from 1D to 100D.
The training data would be the data without missing values; the test data would be the data w/ missing values.
Dear Xu Cui,
Thank you, I found a library for SVM which doesnt require MEX function at all. It process all the SVM functions. However Mr.Xu, Can i know at which place in your coding I should change the single dimension to two dimension. For my case mxn(100×40). So can you please post me a code that shall fit my data? I have clearly understood the meaning of test and train. However, I have only one set of data with missing values. The data(mxn 100×40) is actually, 100 patients answering 40 questions. Its a survey data. In this case, how shall I do it?
I was thinking, I have the data 100×40. Using knn function in Matlab, I will impute the missing values. Next, with the matrix that is imputed, I will create a matrix with missing values. In this way, I can tell that my test data should approximate to training data. Am I right to say that?
In the case of missing values, what number should I represent as? Like “NAN” or something?
Dear Xu Cai,
Can i know your email so that I can send the data, if your email is confidential please test email to I really appreciate for the help you are providing me. It is really helping me. Thank You.
Hi Xu CAi,
I am using LibSVM in regression for training Discrete Wavelet transform coefficients for use in image compression.Let a subband be of size 64×64 (ie: in each column there is a vector of size 64×1). I am training each 64×1 vector with svmtrain and encoding the weights thereby obtained. When I use Libsvm I get a model structure which I may need to pass to decompression routine also. But in realty this should not happen since only the weights should be transmitted and I want to recreate my wavelet coefficients back from these weights. Is there any workaround? This may require lot of code change right? With quadprog (qp solver in matlab) there is no issue.However the computation time is really an issue.
@Bijoy Viswanath
Bijoy,
I am not familiar with image compression. From what you described, I am not sure if you are able to reconstruct the original coefficients from svr weights. The number of weights for a 2D SVM is 2, which I doubt is enough to reconstruct the original data.
But I may not understand your problem enough.
Dear Xu Cui,
I am new to SVR. I have Matlab R2009b version. I am unable to instal LIBSVM package on it. Please help.
Thanks
S ZAIDI
dear mr Xu Cui
I am a beginner in svr and i would like to use svr for 3 input vectors and 1 output vector.I would like to use LIBSVM but i could not use it… i have matlab 7.10.0 R2010a and i put the extracted libsvm in my matlab path and i tried to open make.m and from readme file i tried to change my compiler to Visual C++ but i have not got this choice in my matlab…i have Visual C++ in my laptop but the matlab dose not know it as its compiler…
what can i do for using libsvm and svr???
could you possibly help me???
I am waiting to hearing from you.it would be a kind of you.
best regards
a.b
Type the following command on the matlab prompt.It will find the compiler for you and give you option to select.
Hope it will help you.
mex -setup
hi,
please tell me how to compile the libsvm 3.11
i used the folllowing procedure
1) mex -setup
2) i have selected compiler Microsoft Visual C++ 2005 SP1
3) make
after this command it gives error like this
>> make
LINK : fatal error LNK1181: cannot open input file ‘user32.lib’
C:\PROGRA~1\MATLAB\R2010A\BIN\MEX.PL: Error: Link of ‘libsvmread.mexw32’ failed.
please respond me as early as possible….
Thank you very much for sharing the code. 🙂
Hello
I am an Algerian doctoral student and I work on modeling (artificial intelligence).
If you can give me the MATLAB (Code) program for support vector machine (SVM).
Thank you
Do you know how to scale the test data, based on the min & max of the train data?
Yes. Test and training data should be scaled in the same way.
@lily
Hello there,
I have a question about normalizing… do we have to normalize every column of our date separately respecting to their own min&max data?
for example I have 4 column of input data and 1 column of output data, do I have to normalize every column separately, or normalize the whole data with one min & max?
—————————-
Thanks.
@Hamed
Yes, it’s better to normalize every column individually.
Hi,
Another question,
when I’m using svmtrain, I get this error: “Group must be Vector.”
My input data is not vector. I have 4 column for input data.
and another problem is that when I want to use a vector data for input in another job I get this error: “??? Error using ==> sprintf
Function is not defined for sparse inputs.
Error in ==> num2str at 129
t = sprintf(f,x(i,:));
Error in ==> grp2idx>uniquep at 85
b = cellstr(strjust(num2str(b), ‘left’));
Error in ==> grp2idx at 23
[gn,i,g] = uniquep(s); % b=unique group names
Error in ==> svmtrain at 128
[g,groupString] = grp2idx(groupnames);
Error in ==> libsvmTest at 19
model = svmtrain(Xtest, Xtrain, options);”
I’m looking forward to hearing from you.
Thanks for your time,
Regards
Hi,
why didn’t you normalize “y”?
@Hamed
Good question. I really don’t know why people usually don’t normalize y. If you find out the reason please let me know.
i want to know how to install libsvm in matlab. Please give the step by step procedures as i am beginner.
Hi, I have tested your SVM package for regression,
using matlab 7.10 (R2010a)first there is an error message about normalize, I have used deleted x = normalize(x) and replaced x= mapstd(x), however there is always error message about : Undefined function or method ‘svmtrain’ for input arguments of type ‘double’
why. is possible to give an example to follow it, for regression input/output purpose, of course
thank you
@Tar
Tar, Did you add libsvm into matlab path?
Hi,
I have added path into matlab, like :addpath(‘c:/matlab/libsvm’), but
there is always error message
rror in ==> test_svr at 26
tic;model = svmtrain(y(1:N/2),x(1:N/2,:),[‘-s 4 -t 2 -n ‘ num2str(ii/2) ‘ -c ‘ num2str(1)]);toc
why.
i am new to the support vector regression and using SVR for the prediction of the sales…can you provide me the algorithm of SVR in libsvm….
Hi,
i want to use libsvm on windows. i have downloaded it from the website, extracted it. now what to do next i am not getting please could you explore it step by step. though it is simple question but i am new to this.
Hi,
in svm regression,when we want to prove formula,we have alfa+ and alfa- ,
in libsvm where can i find value of these alfa?
Your Libsvm tutorial is very useful,Thanks for the same.
I am using matlab.
I have 4 columns & 55 rows data (real numbers)how to the scaling for this data & how to convert the data to libsvm format & save in libsvm format,also can we give the testing.txt file in comma separated or tab seperated file.Please let me know your suggestion to do the following
scaling
Coverting to libsvm data format.how to normalise or scale the data to[-1,+1] in matlab.
I am using RBF kernel the accuracy is very low,I have used matlab.
Looking forward for your reply
Regards
syeda
@Syeda
I have updated this post and added normalize function there.
Hi
I want to use your code to find estimate output if training set consists of (x,y) x being the input and y being the output and the test set consists of (x’,y’). Given x’ we have to estimate y’. Kindly help me.
@Navneet
Feel free to use the code.
Hi,
I have data as below:
GENDER Nationality Grade Age (Days) Service (Days)
F Dutch CC.02 10679 789
F South African CC.03 9313 1263
M Brazilian FD.06 17150 1443
F Chinese CC.02 8190 152
M Trinidadian CC.02 9196 722
F Filipino CC.03 10418 2010
F Filipino CC.03 9628 1082
F French CC.04 10556 1950
Hi,
I have data as below: with Service as output vector, Total sample size being 4500
GENDER Nationality Grade Age(days)Servicedays)
F Dutch CC.02 10679 789
F South African CC.03 9313 1263
M Brazilian FD.06 17150 1443
F Chinese CC.02 8190 152
M Trinidadian CC.02 9196 722
F Filipino CC.03 10418 2010
F Filipino CC.03 9628 1082
F French CC.04 10556 1950
Being a new being to SVM, please help me in
–preparing the data, as input vector is not in numerical format
–scaling data
–choosing C,gamma
–predicting
is the code used above can be used for solving?
@Suresh gorakala
Suresh,
For categorical data, you can convert them into numeric values. For example, a three-category attribute such as {red, green, blue} can be represented as (0,0,1), (0,1,0), and (1,0,0).
Scale: linearly scale to range -1 to 1, or 0 to 1
Find best C and gamma, see http://www.alivelearn.net/?p=912
You can use the code above
More info:
http://www.csie.ntu.edu.tw/~cjlin/papers/guide/guide.pdf
@Xu Cui
Thanks Xu Chi,
In My data,if I have 100 Nationalities then do I need to represent in 100 category category attribute?
@Suresh Gorakala
I would do that. Hopefully the number of samples is much larger than 100.
@Xu Cui
Thank you Xu,
Yeah, I the samples are almost 5000 & there are two classes which have categorical attributes, will do as you adviced & will get back to you incase of any issue.
@Xu Cui
Hi Xu,
How to do the normalization between (-1,1) using the below expression:
data = (d -repmat(min(d,[],1),size(d,1),1))*spdiags(1./(max(d,[],1)-min(d,[],1))’,0,size(d,2),size(d,2));
Regards,
Suresh G
@Suresh Gorakala
d is your feature matrix, each row a sample and each column a feature. data is your feature matrix after normalization.
@Xu Cui
—I did understand that but when using the above expression, Normalising is happening in the range of (0,1), but not in the range of (-1,1).
Also could you explain the looping conditions which I’m unable to understand: for M=m ,for ii =1 for JJ=1.
I observe that (for ii=1 & jj=1) does run only once & for M=m runs for 10 times , could you plz explain?
@Suresh Gorakala
I see. Normalization to (0 1) is fine. There is no difference.
ii and jj was supposed to try different parameter values, but not any more (just one time). M is to test for different dimensions of data.
@Xu Cui
Correct me if I’m wrong, for normalizing the test data I need to use the min/max values of the training data in the normalize equation.
i.e
data_test = (d_test -repmat(min(d_train,[],1),size(d_test,1),1))*spdiags(1./(max(d_train,[],1)-min(d_train,[],1))’,0,size(d_test,2),size(d_test,2));
@Suresh Gorakala
You are right. You need to normalize the test and training data in the same way.
@Xu Cui
Thank you Xu for the replies,
Do we need to denormalize the predicted values (zz lables) we get after executing the [zz,acc] = svmpredict(zeros(size(x_test,1),1),x_test,model)?
After we generate a model using training set, how to validate if our results are valid before predicting the data using test data?
@Suresh Gorakala
No.
Usually you split your data to two parts, one for training, and the 2nd for validating. If the accuracy rate in validation is low, then it means the model is not good.
@Xu Cui
Bear with my naive querries Xu,
Please let me know how to calculate the accuracy rate using SVM, I have done validation using Gradient descent but not with SVM.
– correct me if wrong: ‘-v’ option is used in svmtrain method while selecting ‘c,g’ values, once ‘c,g’ are selected we can use the svmtrain method without ‘v’ option.
@Suresh Gorakala
oops,I thought this is SVM.
For SVR, instead of using “accuracy”, you may use correlation between the predicted and actual values.
If you want to do SVR, you always want to use -v option (3 or 4). You don’t need to specify for spmpredict
Correction to my earlier statement: I have validated trainig data using Gradient Descent technique not while using SVM using LIBSVM.Currently I’m currently working on SVM. 🙂
how to find the correlation between the predicted & actual values?
hi
how are u?
my name is hossein
i am a msc student in water resource management
i need a help. if you impossible,
i want to a matlab code same az MatLab code: test_svr.m
with 2 input and 1 out put with SVM regression with libsvm
Thank you in advanced for your consideration and I am looking forward to
hearing from you soon. Yours sincerely, H. Orouji (mail: [email protected])
hi
how are u?
my name is hossein
i am a msc student in water resource management
i need a help. if you impossible,
i want to a matlab code same az MatLab code: test_svr.m
with 2 input and 1 out put with SVM regression with libsvm
Thank you in advanced for your consideration and I am looking forward to
hearing from you soon. Yours sincerely, H. Orouji
@hossein
hi
thanks a lot for reply
how do i create a train and test set with 2 input and 1 output?
i need a code (sample MatLab code)
Thank you in advanced for your consideration and I am looking forward to
hearing from you soon. Yours sincerely, H. Orouji
Dear Xu,
One quick question, how to accomodate missing entries in the input data?
for ex: in my data if few Age & Nationalities entries are missing then how to prepare the data for training?
hi
i need a help. if you impossible, how do i create a train and test set with 2 input and 1 output?
i want to a matlab code same as : ex.m MatLab code.
with 2 input and 1 out put with epsilon regression
Thank you in advanced for your consideration and I am looking forward to
hearing from you soon. Yours sincerely, H. Orouji
code: ex code
N = 30;
u = linspace(0,1,N)’;
y = 1 ./ (0.1 + u) + 0.3*randn(N,1);
svm_type = 3;
kernel_type = 2;
gamma = 10;
cost = 10;
epsilon = 0.2;
options = [‘-s ‘, num2str(svm_type),…
fprintf(‘Starting LIBSVM\n’);
tic;
model = svmtrain(y, u, options);
fprintf(‘Optimization finished in %3.2f sec\n’,toc);
yp = svmpredict(y, u, model);
figure
plot(u,y,’*’)
hold
plot(u,yp,’r’,’linewidth’,2)
xlabel(‘u’)
ylabel(‘y’)
legend(‘process’,’model’)
Hi,
Please advice on how to accommodate missing entries in the input data?
@Suresh Gorakala
Suresh, I myself don’t have experience in handling missing data. I googled and found there is a package called Weka but I have no experience with this package. If you find a solution please let me know.
@Xu Cui
I have done in the following way,
Averaging all the feature (Column) values and replacing the missing values with the resultant average value. This worked for some extent but searching for better ways.
@hossein
hossein, sorry for not replying you earlier. I think to have 2 inputs is fairly easy, simple grouping the two input into one matrix with 2 columns – unless there is something in your question I don’t understand.
Hi! I’m working with precomputed kernel matrix. I guess that svmtrain works well but I’m not sure because the svmpredict results are wrong. Could you give me some help with my code?
sigma = 0.8;
rbfKernel = @(X,Y) exp(- sigma .* DistanceMatrix(X,Y).^2);
k1train = rbfKernel(trainz,trainz);
c = 1;
distance1 = @(X,Y) 1 – DistanceMatrix(X,Y).^2 ./ (DistanceMatrix(X,Y).^2 + c);
k2train = distance1(trainc, trainc);
alpha = 0.6;
k = alpha.*k1train + (1-alpha).*k2train;
ktrain = [(1:ntrain)’, k];
model1 = svmtrain(trainy’, ktrain, ‘-s 3 -t 4 -c 1 -p 0.01′);
k1test = rbfKernel(testz,testz);
k2test = distance1(testc, testc);
ktest = (alpha)*k1test + (1-alpha)*k2test;
kt = [(1:neval)’, ktest];
yfit3 = svmpredict(testy’, kt, model1)
I am working with libsvm for regression model. I would like to know how support vectors can be identified in the final model structure. I know determining the number of support vectors in regression problem. However, I dont not know which patterns from training set are SV. Thank you
@maria
maria,
Sorry I don’t know the answer either (I only did it for SVM case). If you find out, please let me know.
@satish
Were you able to solve this error? I’m getting the exact same error, and there is no other help available on the net.
Dear Xu Cui,
I am trying to use LibSVM for travel time prediction. I have a feature vector matrix of form (latitude_StartPoint, longitude1_StartPoint, latitude2_EndPoint,longitude_EndPoint, distancebt_points,…….) and training_label_vector as time_taken_bt_points. My doubt is do I need to normalize both input and output vector
@chanukya
As long as your features are in similar range, it should be fine not normalize. I think you can try both to see if there is any real difference.
Thanks Xu Cui for getting back to me. I have one more. I have 50 more instances that I use to train. But every instance has different (latitude1_StartPoint, longitude1_StartPoint, latitude2_EndPoint,longitude_EndPoint) even though I they belong to same route due to floating gps points. So do I need to standardize all the starting and ending points to fixed points(Latitude and Longitude) before I train them or can I proceed with out standardizing to fixed points?
@chanukya
The best way to find out the answer is, I think, to try both ways and see if there is big difference. I suspect there won’t be real difference here.
Thanks Xu Cui,
Did you work on R. Gunn toolbox for regression in matlab. Before I started using LibSVM I tried using that which gave me more accurate results with the same input vector epsilon svr. kernel = ‘linear’;C = 1000;loss = ‘quadratic’;e = 0.001; LibSVM linear kernel[-s 3 -t 0 -n 1:100 -c 1:10].Can you please give me some idea if I am inputing some wrong parameters.
Dear Xu Cui,
I am trying to use epsilon svr with radial basis function. I am trying to use [-s 3 -t 2 -g x -c y -p x]. How can I find the best values of [g c p]. I tried fixing c values and running two loops for g and p to find best values of correlation coefficient. But I am not sure how this works. Even I tried fixing p value and running for c and g. Do LibSVM internally perform cross validation or do I need to run it manually? Can you help me to get some clear idea.
@Aj
No, i did not get any solution on this error…
hi
am new to work with SVM regression.
actually am doing project in image compression using wavelets along with support vector machines . i want to know how to give the coefficients of the image which is obtained from wavelet decomposition as an input to svm regression to get support vectors?
can you please help me
hello sir,
i want to know whether LIBSVM can useful for multi dimension regression (both linear and non linear).
the meaning of 1D regression is to find only “W” and “b”. and meaning of multi dimension regression is there will be more than one “w”. am i correct ?.
vishal mishra
@vishal mishra
libsvm can be used in multi-dimension. There will be mutiple weights. But it is not regression.
@satish
hi Aj
I installed one of the C compiler and created MEX files. Initially it wasnt happening in my PC i dont know why..i did it on my friends PC and created it and just copied MEX files in my folder and it works..
a href=”#comment-1641″>@Xu Cui
thanks sir ………
LIBSVM for matlab interface is written in C/C++ format. is there any tool box or other LIBSVM which is completely written in MATLAB ?.
I think MatLab comes with its own svm program in the bioinfo package.
> which svmtrain
C:\MATLAB\R2012\toolbox\bioinfo\biolearning\svmtrain.m
See:
http://www.mathworks.com/help/stats/svmtrain.html
Hi,
I would like to know how to plot in matlab the hyperplan found for the case of the e-svr with a polymonial (for example ) kernel with 1D feature and not only the red test points (like in your figure above?
thanks
What if if we have only one feature? I mean we have the y label, and want to predict the future days. Which values will we use for x? Is ıt possible to use time (for i.e. days)?
@hakan
I don’t quite understand the question. 1 feature is totally fine.
I’m not sure how to use SVR in either univariate and multivariate time series.
Say we have stock prices for N days. For training inputs, y are the stock prices for N days, but what will we use for x ?
1.Time series? For i.e. in one step ahead prediction 1,2,3…Z for Z days?
2.(for one step ahead) sifting one day of y values?
To explain more:
matlab> model = svmtrain(training_label_vector,
training_instance_matrix [, ‘libsvm_options’]);
For univariate: I use the stock prices for N days in training_label_vector as a column vector and want to predict say next 30 days. I wonder which data I have to use in training_instance_matrix?
For multivariate: say I have 22 more features (prices of other goodies), I use other features as column vectors in training_instance_matrix. But I’m not sure if I’m using the correct approach.
@hakan
You can use earlier stock price as X. For example, Y is the stock price at day 2, 3, …, N, X can be the stock price at day 1, 2, 3, …, N-1
Hi,
I have been using LIBSVM for along time. A regression problem is my interest. My problem is very important and i couldn’t solve it. I have two inputs and one output for SVR. Firstly i normalize my train data (both inputs and target) between 0 to 1 or -1 to 1 individually, and train the SVR with this normalized data, After training i use a test set which i similarly normalize test set by using min/max values of training stage. Finally i get the predicted values between 0 – 1 or -1 – 1. But when i denormalize this predicted values to actual values by using max/min values of target in training satage i get a prediction set which can not reach the max and min values of actual data. For axample in test stage actual outputs change between 0 – 100 but the predicted values change between 10 – 90. I tried lots of ways but couldn’t able to salve this. Please help me,
Thanks
Hello Xu Cui,
I have question regarding selection of input features. Could you please look into the link where I posted my question http://stackoverflow.com/questions/18242784/training-libsvm-with-multivariate-data-in-matlab
Could you please tell me which features should I select for training and testing? Please look into comments for the second response, I have given more description of my problem.
For a time series data how can I input the time. I have my data collected on 6 different days at times starting from 8:00 to 11:00 am.So how do I input this data? Initially I get the UNIX time-stamp which I converted into day, time and entered into 2 different columns. For example now I have variable 1 as day with values 12,13,14,15,16,17 and variable 2 with times 8.0,8.25,8.5,8.75,9.0,9.25,…..11.0,8.0,8.25,8.5,8.75,9.0,9.25,…..11.0,8.0,8.25,8.5,8.75,9.0,9.25,…..11.0,8.0,8.25,8.5,8.75,9.0,9.25,…..11.0,…………….
I have the data collected with 15minutes of frequency, so the experiment has all the stuff repeated for 6 days.
So now I would be glad if you can suggest me how I could train LIBSVM with this input time series.
@serkan
serkan mail me [email protected]
Hello Sir,
I have developed svm_model for regression, now i want to test it for a given input’x’. after reading the readme file i got that i should use
Function: double svm_predict(const struct svm_model *model,
const struct svm_node *x);
but i do not know how to use? please help so that i can use it in matlab.
thanks and regards
Vishal mishra
hi my friend
i need your honestly help, i dont know what does svmtrain and quadprog in matlab mean?
i dont know how should i solve this question?
Suppose we have 5 points in 1D feature space as follows:
x1=1, x2=2, x3=4, x4=5, x5=6, with 1, 2, 6 as class 1 and 4, 5 as class 2
Use a polynomial kernel of degree 2 as K(x,z) = (xz+1)2 to find
To do that, follow these steps:
1) Find αi (i=1, …, 5) by
2) Find support vectors
3) Find the discriminant function f(z)
4) Find the bias b
5) Plot the discriminate function, 5 points and the regions for the classes
Note that y=1 if x in w1 and y=-1 if x in w2
@Yang Fei
check your copy/paster ‘ character in normalize 😉
Best.
Sir,
My proj deals with emotion recognition from speech. Given that I have 40 samples of 6 emotions each, I have extracted the feature of mfcc from each of the sample. However, because the length of each speech sample is different, the order of the extracted mfcc’s are also different. does svm deal with unbalanced sizes of data? also, in the svmtrain and svmclassify programs available in the biolearning toolbox of matlab, do we need to add any parameters?
Hi,
Can you please send me the MATLAB code for Nonlinear function approximation.(Some thin like above parabolic function)
Sir,
While running LIBSVM – Multiclassification in MAT lab i’m getting the following error
??? Error using ==> svmtrain at 172
Group must be a vector.
Error in ==> ovrtrain at 8
models{i} = svmtrain(double(y == labelSet(i)), x, cmd);
Error in ==> get_cv_ac at 9
model = ovrtrain(y(train_ind),x(train_ind,:),param);
Error in ==> automaticParameterSelection at 67
cv = get_cv_ac(trainLabel, trainData, cmd, Ncv);
Error in ==> sample at 88
[bestc, bestg, bestcv] = automaticParameterSelection(evalLabel, evalData, Ncv_param, optionCV);
To make it work, make sure that the LibSVM library is part of your Matlab’s Search Path. One option would be to use the Matlab filebrowser (Current Folder) to go the LibSVM folder and use the menu Add to Path -> Selected Folders and Subfolders. _____________ I have tried this solution also.
But i couldnt rectify my problem. Kindly help me to sort out this problem.
Regards,
Angeline
could you please send the matlab code for the followin raw data and show hw to scale it between [0,1] and[+1,-1]
function data = normalize(d)
% scale before svm
% the data is normalized so that max is 1, and min is 0
data = (d -repmat(min(d,[],1),size(d,1),1))*spdiags(1./(max(d,[],1)-min(d,[],1))’,0,size(d,2),size(d,2));
please could u tell how to accomodate(load) this data and perform scaling
and call svmtrain and classify
please send the matlab code for the same
4 columns(feartures) and 55 samples(columns)
7 classess
4 features
for ex:
1 10 20 30 40
1 values
1 -do-
1
1
1
1
2
2
2
2
2
2
2
3
3
3
3
3
3
3
3
4
4
4
4
4
4
4
4
5
5
5
5
5
5
5
5
6
6
6
6
6
6
6
6
7
7
7
7
7
7
7
7
looking forward for ur reply..
Hello Xu Cui,
I am working in EEG signal processing field. I need nonlinear SVM for classifying my EEG signals.Can you please share the SVMtrain and svmclassify codes in matlab.
Hello Xu Cui,
I have done feature extraction of EEG signal(for epilepsy localization)using ICA. And got topoplot(in MATLAB) of independent components.Now i need to generate a matrix from this topoplot to give as an input for classification of signal(using SVM).How we can gennerate the matrix?
I need your help to develop svm model using regression, how do i create a train and test set with 4 input column and 1 output column?
Hello sir
please help me .I have already intimated you about my problem.give some idea so that i can do my research work.
Hi,
what happens if the normalization has values inside [-5,5] or [-1,1]. [0,1] is loosing information ?
@Andre
Scaling to [0,1] is fastest and does Not lose you information. It could be that the SVM predicts slightly different prediction values depending on how you do your scaling. Remember to scale prediction date in the same way also.
Hi,
I’d like to use this library for outputs with two values, for example:
X = [[2,5,3,8,1,1,1],[2,3,3,8,0,0,1],[3,5,1,2,1,1,1],[2,5,4,8,5,1,1]]
y = [(2,3),(5,8),(5,7),(7,0)]
I had an error when i tried this about the length of the output.
Thanks
@Char_lee
I don’t think svr handles 2D output
hi, Xu,
I have experienced similar experience as the author of of the following post.
please advise any fallacy of the code. thanks
http://stackoverflow.com/questions/18300270/lag-in-time-series-regression-using-libsvm
@Ryan Lam
Very interesting phenomenon. see
http://www.alivelearn.net/?p=1666
Respected Sir,
I am new to SVM. My feature set is of 100×24 (random number between 10:100) and my label is 100×1 (random numbers between 10:100). But let us say it has 5 class and I have to use svm-scale to normalize the data. How I have to do it?
I WANT INFORMATION ABOUT THE .range4 FILE and svmguide4 IN THE SVM-SCALE (below syntax is defined).
How it is created?, IF I want to create it how should I create it.
PLEASE EXPLAIN IT WITH SMALL EXAMPLE
../svm-scale -l 0 -s range4 svmguide4 > svmguide4.scale
for example (JUST randomly I have given)
X 1 -1
1 9 20
2 56 98
…
Thanking you sir
I am using libsvm for regression problem How to check acuracy cross validation of C, gamma and epsilon parameters? Is it cross validation accuracy or Mean Square Error?
Respected Sir
There is any option for methodological change in SVM. Can we update the Kernal of SVM using any clustering technique.THen kindly explain how it be
I would like to apply SVM and one more clustering technique.
Thank u sir
whether it is possible to predict future values using SVR?
I have some question Sir,
How much the number of support vector generated by SVR in a good case? is that normal if i got 90 SV from 100 data ?
And is there any rule of minimum training data to build SVR model ?
Hello sir
I am using libsvm for regression. the Input is processed signal EMG and the output is measured torque. I want to use the epsilon-SVR to estimate knee torque by EMG test signal, but the estimated output is zero!
number of samples (EMG & Torqe) for train is 801*1
number of samples (EMG & Torqe) for test is 310*1
and all of them is normalised between 0,1 (by samples/max(samples))
and My code is:
model = svmtrain(Torqetrain,EMGtrain,’-s 3 -c 40 -t 2 -p 0.1 -d 5′);
[PredictedY,MSE] = svmpredict(Torqetest,EMGtest,model);
my model after train is:
model.Parameters [3;2;5;1;0]
model.nr_class 2
model.totalSV 281
model.rho -2.521
model.Label [ ]
model.sv_indices 281×1 double
model.ProbA [ ]
model.ProbB [ ]
model.nSV [ ]
model.sv_coef 281×1 double
model.SVs 281×1 sparse double
can you help me please?
Normalizing data range to [0,1] may not be enough. Train data divided by their standard deviation (std) may lead to better results.
If so, you have to divide test data by the constant value given by std previously to be consistent.
Hi,
I am new to SVR, and would like to try your program, test_svr.m, before starting with my own data set. I can’t locate your data file.
I also clicked your tab: “Download link sent”. I didn’t get the email. Checked spam version as well. Nothing there either. Could you please send me the link to the e-mail address shown above?
Thanks.
Hi again.
I proceeded with your program using the random number generator. Now it is having problem with corrcoef function. Do I need to download some other program?
Dear Mr Cui, I am new in matlab and SVM coding. Please I want to know how I can obtain the frequency, Scale, Phase etc parameters from Matching Pursuit and Gammatonne Frequency Cepstral Coefficient algorithms in order to feed them into the SVM classifier to detect and recognize environmental sound. Thank you very much as I await your response
Dear Cui
Please how can I get the mean and variance of each my GFCC frame coefficient, so that I can feed it to my SVM classifier.
Thank you
Again, please I do I show that a particular frame is an environmental sound frame or not. that is what are the criteria to know that a frame is an environmental sound frame. Thank you again
Please, still waiting for your reply
down vote
favorite
i’m going to use the SVM with the iris data when i calculate the kernel function ,i have an error i’can’t fixed .some help please
model1 = svmtrain( LS’,SSapp’,’-s 1 -t 1 -g 0.25 -c 0 -n 0.5 -r 0.35 -d 1 -b 1′); Error in svmtrain (line 57) errstring = consist(net, ‘svm’, X, Y);
Error in irissataset (line 23) model1 = svmtrain( LS’,SSapp’,’-s 1 -t 1 -g 0.25 -c 0 -n 0.5 -r 0.35 -d 1 -b 1′);
How can I use the probability values instead of labels in training data using svm regression?