recon-all is a batch program and runs >30 steps. It easily takes 30 hours to finish one subject.
Use tkmedit CON14 T1.mgz and tksurfer CON14 lh inflated to visualize images. More about visualization with freesurfer.
Note: commands are in green. They are copied from the log files.
View images for every step here
Before you start, you need to put the structural images into certain directory hierarchy and set environment variable. Assume folder “structural” is where subjects’ structural images are. Under “structural”, you have folders “SUBJ1”, “SUBJ2”, “CON14” etc for every subject. You will have to put your structural images to “structural/SUBJ1/mri/orig/001.mgz”. If you have multiple images for this subject, use “002.mgz” etc. (If you original file format is not mgz, check out how to convert image formats).
structural
|--SUBJ1
|--mri
|--orig
|--001.mgz
|--002.mgz
Then set environment variable in linux shell:
setenv SUBJECTS_DIR $PWD
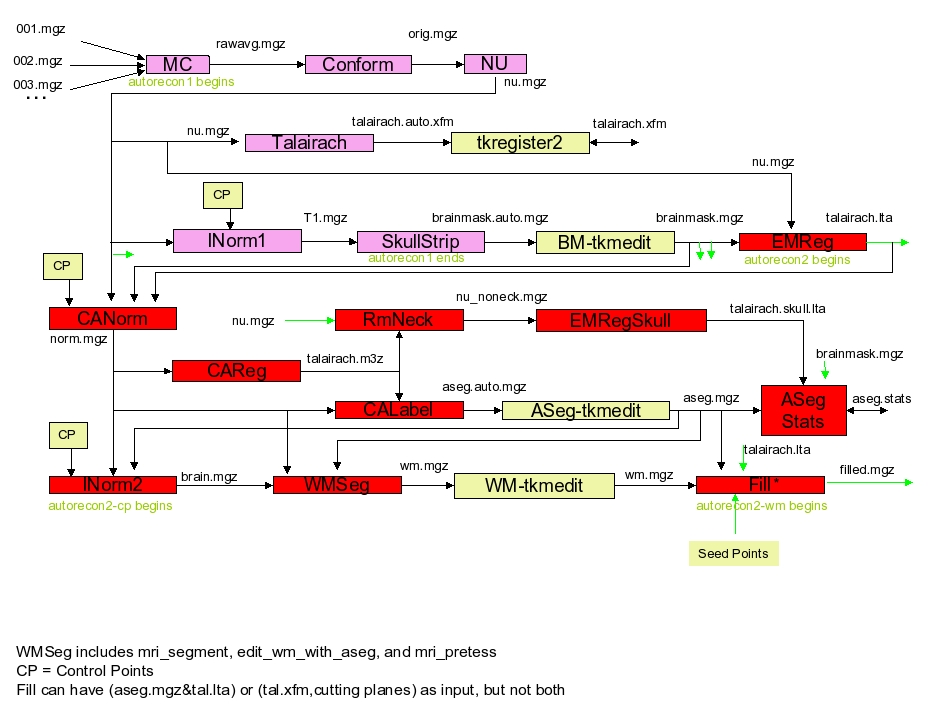
- motion correction, and conform
input: mri/orig/00?.mgz
output: mri/orig.mgz (through mri/rawavg.mgz)
If you have multiple structural images (001.mgz, 002.mgz, etc), then motion correction will be done and rawavg.mgz is created. If you have only one image (001.mgz), it simply copies the file and rename to rawavg.mgz.
cp CON14/mri/orig/001.mgz CON14/mri/rawavg.mgz
Then conform using mri_convert:
mri_convert CON14/mri/rawavg.mgz CON14/mri/orig.mgz --conform
Conform is to make voxel size 1x1x1mm and same number of voxels (256) along x,y,z directions (iso). Data is written to orig.mgz. - non-uniform intensity normalization
input: mri/orig.mgz
output: mri/nu.mgz
mri_nu_correct.mni --i orig.mgz --o nu.mgz --n 2
Non-parametric Non-uniform intensity Normalization (N3), corrects for intensity non-uniformity in MR data, making relatively few assumptions about the data. This runs the MINC tool ‘nu_correct’. By default, four iterations of nu_correct are run. The flag -nuiterations specification of some other number of iterations. - talairach transformation
input: mri/nu.mgz
output: transform/talairach.auto.xfm
talairach_avi --i nu.mgz --xfm transforms/talairach.auto.xfm
This computes the affine transform from the orig volume to the MNI305 atlas using the MINC program mritotal (see Collins, et al, 1994) through a FreeSurfer script called talairach. Several of the downstream programs use talairach coordinates as seed points. You can/should check how good the talairach registration is usingtkregister2 --s subjid --fstal. tkregister2 allows you to compare the orig volume against the talairach volume resampled into the orig space. Run “tkregister2 –help” for more information. Creates the files mri/transform/talairach.auto.xfm and talairach.xfm.
This step produces a long log. - intensity normalization
input: mri/nu.mgz
output: mri/T1.mgz
mri_normalize -g 1 nu.mgz T1.mgz
Performs intensity normalization of the orig volume and places the result in mri/T1.mgz. Attempts to correct for fluctuations in intensity that would otherwise make intensity-based segmentation much more difficult. Intensities for all voxels are scaled so that the mean intensity of the white matter is 110. If there are problems with the normalization, users can add control points. See also Normalization2. - skull strip
input: mri/nu.mgz
output: mri/brainmask.mgz, mri/brainmask.auto.mgz
mri_em_register -skull nu.mgz /usr/local/freesurfer/average/RB_all_withskull_2008-03-26.gca transforms/talairach_with_skull.lta
Removes the skull from mri/T1.mgz and stores the result in mri/brainmask.auto.mgz and mri/brainmask.mgz. Runs the mri_watershed program. If the strip fails, users can specify seed points (-wsseed) or change the threshold (-wsthresh, -wsmore, -wsless). The -autorecon1 stage ends here. - EM (GCA) Registration (-<no>gcareg)
input: mri/nu.mgz, mri/brainmask.mgz
output: mri/transforms/talairach.lta
mri_em_register -mask brainmask.mgz nu.mgz /usr/local/freesurfer/average/RB_all_2008-03-26.gca transforms/talairach.lta
Computes transform to align the mri/nu.mgz volume to the default GCA atlas found in FREESURFER_HOME/average (see -gca flag for more info). Creates the file mri/transforms/talairach.lta. The -autorecon2 stage starts here. - CA Normalize (-<no>canorm)
input: mri/nu.mgz, mri/brainmask.mgz
output: mri/norm.mgzmri_ca_normalize -mask brainmask.mgz nu.mgz /usr/local/freesurfer/average/RB_all_2008-03-26.gca transforms/talairach.lta norm.mgz
Further normalization, based on GCA model. Creates mri/norm.mgz. - CA Register (-<no>careg)
input: mri/norm.mgz, mri/brainmask.mgz
output: talairach.m3z
mri_ca_register -align-after -nobigventricles -mask brainmask.mgz -T transforms/talairach.lta norm.mgz /usr/local/freesurfer/average/RB_all_2008-03-26.gca transforms/talairach.m3z
Computes a nonlinear transform to align with GCA atlas. Creates the file mri/transform/talairach.m3z.
This step is very loooooong (10 hours) - Remove neck (-<no>rmneck)
input: mri/nu.mgz
output: mri/nu_noneck.mgz
mri_remove_neck -radius 25 nu.mgz transforms/talairach.m3z /usr/local/freesurfer/average/RB_all_2008-03-26.gca nu_noneck.mgz
The neck region is removed from the NU-corrected volume mri/nu.mgz. Makes use of transform computed from prior CA Register stage. Creates the file mri/nu_noneck.mgz. - EM Registration, with Skull (-<no>skull-lta)
input: mri/nu_noneck.mgz
output: mri/transforms/talairach_with_skull.lta
mri_em_register -skull -t transforms/talairach.lta nu_noneck.mgz /usr/local/freesurfer/average/RB_all_withskull_2008-03-26.gca transforms/talairach_with_skull.lta
Computes transform to align volume mri/nu_noneck.mgz with GCA volume possessing the skull. Creates the file mri/transforms/talairach_with_skull.lta. - CA Label (-<no>calabel)
input: mri/norm.mgz, mri/transforms/talairach.m3z
output: mri/aseg.auto.mgz, mri/aseg.mgz, (through mri/aseg.auto_noCCseg.mgz)
mri_ca_label -align -nobigventricles norm.mgz transforms/talairach.m3z /usr/local/freesurfer/average/RB_all_2008-03-26.gca aseg.auto_noCCseg.mgz
mri_cc -aseg aseg.auto_noCCseg.mgz -o aseg.auto.mgz CON14
Labels subcortical structures, based in GCA model. Creates the files mri/aseg.auto.mgz and mri/aseg.mgz. - ASeg Stats (-<no>segstats)
?Computes statistics on the segmented subcortical structures found in mri/aseg.mgz. Writes output to file stats/aseg.stats. - Normalization2 (-<no>normalization)
input: mri/norm.mgz, mri/aseg.mgz, mri/brainmask.mgz
output: mri/brain.mgz
mri_normalize -aseg aseg.mgz -mask brainmask.mgz norm.mgz brain.mgz
mri_mask -T 5 brain.mgz brainmask.mgz brain.finalsurfs.mgz
Performs a second (major) intensity correction using only the brain volume as the input (so that it has to be done after the skull strip). Intensity normalization works better when the skull has been removed. Creates a new brain.mgz volume. The -autorecon2-cp stage begins here. If -noaseg flag is used, then aseg.mgz is not used by mri_normalize. - WM Segmentation (-<no>segmentation)
input: brain.mgz
output: wm.seg.mgz
mri_segment brain.mgz wm.seg.mgz
mri_edit_wm_with_aseg -keep-in wm.seg.mgz brain.mgz aseg.mgz wm.asegedit.mgz
mri_pretess wm.asegedit.mgz wm norm.mgz wm.mgz
Attempts to separate white matter from everything else. The input is mri/brain.mgz, and the output is mri/wm.mgz. Uses intensity, neighborhood, and smoothness constraints. This is the volume that is edited when manually fixing defects. Calls mri_segment, mri_edit_wm_with_aseg, and mri_pretess. To keep previous edits, run with -keepwmedits. If -noaseg is used, them mri_edit_wm_aseg is skipped. - Cut/Fill (-<no>fill)
input: wm.mgz
output: filled.mgz
mri_fill -a ../scripts/ponscc.cut.log -xform transforms/talairach.lta -segmentation aseg.auto_noCCseg.mgz wm.mgz filled.mgz
This creates the subcortical mass from which the orig surface is created. The mid brain is cut from the cerebrum, and the hemispheres are cut from each other. The left hemisphere is binarized to 255. The right hemisphere is binarized to 127. The input is mri/wm.mgz and the output is mri/filled.mgz. Calls mri_fill. If the cut fails, then seed points can be supplied (see -cc-crs, -pons-crs, -lh-crs, -rh-crs). The actual points used for the cutting planes in the corpus callosum and pons can be found in scripts/ponscc.cut.log. The stage -autorecon2-wm begins here. This is the last stage of volumetric processing. If -noaseg is used, then aseg.mgz is not used by mri_fill. - Tessellation (-<no>tessellate)
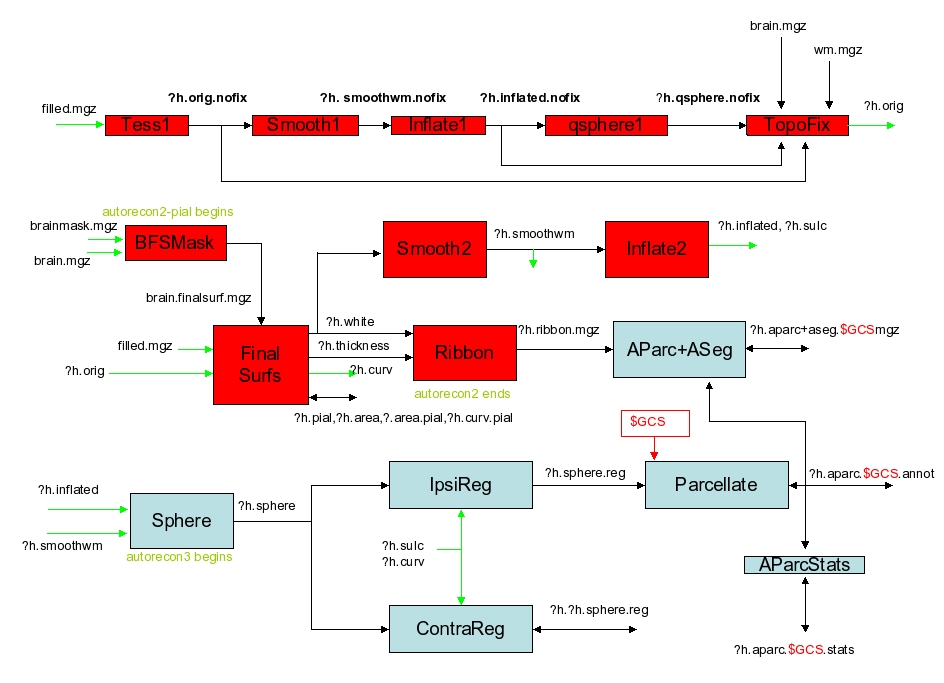
input: filled.mgz, norm.mgz
output: surf/lh.orig.nofix (through filled-pretess255.mgz)
mri_pretess ../mri/filled.mgz 255 ../mri/norm.mgz ../mri/filled-pretess255.mgz
mri_tessellate ../mri/filled-pretess255.mgz 255 ../surf/lh.orig.nofix
mris_extract_main_component ../surf/lh.orig.nofix ../surf/lh.orig.nofix
This is the step where the orig surface (ie, surf/?h.orig.nofix) is created. The surface is created by covering the filled hemisphere with triangles. Runs mri_tessellate. The places where the points of the triangles meet are called vertices. Creates the file surf/?h.orig.nofix Note: the topology fixer will create the surface ?h.orig. - Orig Surface Smoothing (-<no>smooth1, -<no>smooth2)
input: surf/lh.orig.nofix
output: surf/lh.smoothwm.nofix
mris_smooth -nw ../surf/lh.orig.nofix ../surf/lh.smoothwm.nofix
After tesselation, the orig surface is very jagged because each triangle is on the edge of a voxel face and so are at right angles to each other. The vertex positions are adjusted slightly here to reduce the angle. This is only necessary for the inflation processes. Creates surf/?h.smoothwm(.nofix). Calls mris_smooth. Smooth1 is the step just after tessellation, and smooth2 is the step just after topology fixing. - Inflation (-<no>inflate1, -<no>inflate2)
input: surf/lh.smoothwm.nofix
output: surf/lh.inflated.nofix, lh.sulc, lh.curv, lh.area
mris_inflate -no-save-sulc ../surf/lh.smoothwm.nofix ../surf/lh.inflated.nofix
Inflation of the surf/?h.smoothwm(.nofix) surface to create surf/?h.inflated. The inflation attempts to minimize metric distortion so that distances and areas are perserved (ie, the surface is not stretched). In this sense, it is like inflating a paper bag and not a balloon. Inflate1 is the step just after tessellation, and inflate2 is the step just after topology fixing. Calls mris_inflate. Creates ?h.inflated, ?h.sulc, ?h.curv, and ?h.area. - QSphere (-<no>qsphere)
input: lh.inflated.nofix
output: lh.qsphere.nofix
mris_sphere -q ../surf/lh.inflated.nofix ../surf/lh.qsphere.nofix
This is the initial step of automatic topology fixing. It is a quasi-homeomorphic spherical transformation of the inflated surface designed to localize topological defects for the subsequent automatic topology fixer. Calls mris_sphere. Creates surf/?h.qsphere.nofix. - Automatic Topology Fixer (-<no>fix)
input: qsphere.nofix
output: lh.orig
mris_fix_topology -mgz -sphere qsphere.nofix -ga CON14 lh
mris_euler_number ../surf/lh.orig
mris_remove_intersection ../surf/lh.orig ../surf/lh.orig
Finds topological defects (ie, holes in a filled hemisphere) using surf/?h.qsphere.nofix, and changes the orig surface (surf/?h.orig.nofix) to remove the defects. Changes the number of vertices. All the defects will be removed, but the user should check the orig surface in the volume to make sure that it looks appropriate. Calls mris_fix_topology. Creates surf/?h.orig (by iteratively fixing surf/?h.orig.nofix). - Final Surfaces (-<no>finalsurfs)
input: lh.orig
output: lh.white, lh.pial, lh.thickness, lh.curv
mris_make_surfaces -noaparc -mgz -T1 brain.finalsurfs CON14 lh
Creates the ?h.white and ?h.pial surfaces as well as the thickness file (?h.thickness) and curvature file (?h.curv). The white surface is created by “nudging” the orig surface so that it closely follows the white-gray intensity gradient as found in the T1 volume. The pial surface is created by expanding the white surface so that it closely follows the gray-CSF intensity gradient as found in the T1 volume. Calls mris_make_surfaces. - [need to double check, b/c in the log this step is not here, also find command such as
mris_calc -o lh.area.mid lh.area add lh.area.pial
mris_calc -o lh.area.mid lh.area.mid div 2
mris_calc -o lh.volume lh.area.mid mul lh.thickness] Cortical Ribbon Mask (-<no>cortribbon)
Creates binary volume masks of the cortical ribbon, ie, each voxel is either a 1 or 0 depending upon whether it falls in the ribbon or not. Saved as ?h.ribbon.mgz. Uses mgz regardless of whether the -mgz option is used. The -autorecon2 stage ends here. - Repeat step 17
output: lh.smoothwm
mris_smooth -n 3 -nw ../surf/lh.white ../surf/lh.smoothwm - Repeat step 18
input: lh.smoothwm
output: lh.inflated
mris_inflate ../surf/lh.smoothwm ../surf/lh.inflated
mris_curvature -thresh .999 -n -a 5 -w -distances 10 10 ../surf/lh.inflated - Spherical Inflation (-<no>sphere)
input: lh.inflated
output: lh.sphere
mris_sphere ../surf/lh.inflated ../surf/lh.sphere
Inflates the orig surface into a sphere while minimizing metric distortion. This step is necessary in order to register the surface to the spherical atlas. (also known as the spherical morph). Calls mris_sphere. Creates surf/?h.sphere. The -autorecon3 stage begins here. - Ipsilateral Surface Registation (Spherical Morph) (-<no>surfreg)
input: lh.sphere
output: lh.sphere.reg
mris_register -curv ../surf/lh.sphere /usr/local/freesurfer/average/lh.average.curvature.filled.buckner40.tif ../surf/lh.sphere.reg
Registers the orig surface to the spherical atlas through surf/?h.sphere. The surfaces are first coarsely registered by aligning the large scale folding patterns found in ?h.sulc and then fine tuned using the small-scale patterns as in ?h.curv. Calls mris_register. Creates surf/?h.sphere.reg. - [can’t find this step, but find command mris_jacobian ../surf/lh.white ../surf/lh.sphere.reg ../surf/lh.jacobian_white ]Contralateral Surface Registation (Spherical Morph) (-<no>contasurfreg)
Same as ipsilateral but registers to the contralateral atlas. Creates lh.rh.sphere.reg and rh.lh.sphere.reg. - Average Curvature (-<no>avgcurv)
input: lh.sphere.reg
output: lh.avg_curv
mrisp_paint -a 5 /usr/local/freesurfer/average/lh.average.curvature.filled.buckner40.tif#6 ../surf/lh.sphere.reg ../surf/lh.avg_curv
Resamples the average curvature from the atlas to that of the subject. Allows the user to display activity on the surface of an individual with the folding pattern (ie, anatomy) of a group. Calls mrisp_paint. Creates surf/?h.avg_curv. - Cortical Parcellation (-<no>cortparc, -<no>cortparc2)
input: mri/aseg.mgz, surf/lh.sphere.reg
output: label/lh.aparc.annot
mris_ca_label -aseg ../mri/aseg.mgz CON14 lh ../surf/lh.sphere.reg /usr/local/freesurfer/average/lh.curvature.buckner40.filled.desikan_killiany.2007-06-20.gcs ../label/lh.aparc.annot
Assigns a neuroanatomical label to each location on the cortical surface. Incorporates both geometric information derived from the cortical model (sulcus and curvature), and neuroanatomical convention. Calls mris_ca_label. -cortparc creates label/?h.aparc.annot, and -cortparc2 creates /label/?h.aparc.a2005s.annot. - Parcellation Statistics (-<no>parcstats)
input: stats/lh.aparc.stats, label/lh.aparc.annot
output: label/aparc.annot.ctab
mris_anatomical_stats -mgz -f ../stats/lh.aparc.stats -b -a ../label/lh.aparc.annot -c ../label/aparc.annot.ctab CON14 lh
Runs mris_anatomical_stats to create a summary table of cortical parcellation statistics for each structure, including 1. structure name 2. number of vertices 3. total surface area (mm^2) 4. total gray matter volume (mm^3) 5. average cortical thickness (mm) 6. standard error of cortical thicknessr (mm) 7. integrated rectified mean curvature 8. integrated rectified Gaussian curvature 9. folding index 10. intrinsic curvature index. For -parcstats, the file is saved in stats/?h.aparc.stats. For -parcstats2, the file is saved in stats/?h.aparc.a2005s.stats. - Repeat step 29 (Cortical Parcellation)
mris_ca_label -aseg ../mri/aseg.mgz CON14 lh ../surf/lh.sphere.reg /usr/local/freesurfer/average/lh.atlas2005_simple.gcs ../label/lh.aparc.a2005s.annot - Repeat step 30 (Parcellation statistics)
mris_anatomical_stats -mgz -f ../stats/lh.aparc.a2005s.stats -b -a ../label/lh.aparc.a2005s.annot -c ../label/aparc.annot.a2005s.ctab CON14 lh - Repeat step 16-32 for right hemisphere
- ASeg Stats
mri_segstats –seg mri/aseg.mgz –sum stats/aseg.stats –pv mri/norm.mgz –excludeid 0 –brain-vol-from-seg –brainmask mri/brainmask.mgz –in mri/norm.mgz –in-intensity-name norm –in-intensity-units MR –etiv –subject CON14 –surf-wm-vol –ctab /usr/local/freesurfer/ASegStatsLUT.txt - Cortical ribbon mask
mris_volmask –label_left_white 2 –label_left_ribbon 3 –label_right_white 41 –label_right_ribbon 42 –save_ribbon –save_distance CON14 - AParc-to-ASeg
mri_aparc2aseg –s CON14 –volmask
mri_aparc2aseg –s CON14 –volmask –a2005s - WMParc
mri_aparc2aseg –s CON14 –labelwm –hypo-as-wm –rip-unknown –volmask –o mri/wmparc.mgz –ctxseg aparc+aseg.mgz - mri_segstats –seg mri/wmparc.mgz –sum stats/wmparc.stats –pv mri/norm.mgz –excludeid 0 –brain-vol-from-seg –brainmask mri/brainmask.mgz –in mri/norm.mgz –in-intensity-name norm –in-intensity-units MR –etiv –subject CON14 –surf-wm-vol –ctab /usr/local/freesurfer/
Check out recon-all manual in freesurfer wiki.



Hi,
my name is Martina and I’m a student of biomedical engineering in Rome. I’m doing a brain imaging research for my final exam.
Thank you for your useful info on your website.
I’m using freesurfer for registration and segmentation of brain images instead of FSL tools (flirt,bet and fast) and then continue to process the data with FSL. But I have a problem: after converting the image generated with freesurfer mri_convert to “.nii” format it does not overlap with FSL’s reference MNI152.
I know that freesurfer uses talairach reference. Is it true? May it be the problem?
How can I superimpose properly the images?
Do you know any workaround?
Thank you very much.
Regards
Martina Andellini
thank you very much, this is very helpful for me
Dear Xu,
Thanks for the explanation, would it be possible to help me on the following?
I want to get the segmented cortex from the Talairach space to the standard space and convert it to a mask so that I can use it to extract the cortex from other scans (T2 and T2*) which have already been co-registered to the original T1 image in its standard space.
Many thanks
Ali